

Statistical methods for analytical method comparison and validation studies. The package implements Bland-Altman analysis, Passing-Bablok regression, Deming regression, precision experiments, and quality goal specifications based on biological variation — approaches commonly used in clinical laboratory method validation.
Install valytics from CRAN:
install.packages("valytics")Or install the development version from GitHub:
# install.packages("pak")
pak::pak("marcellogr/valytics")valytics provides tools for analytical method
validation:
Method Comparison - Bland-Altman analysis: Assess agreement through bias and limits of agreement - Passing-Bablok regression: Non-parametric regression robust to outliers - Deming regression: Errors-in-variables regression for method comparison
Precision Experiments - Variance component analysis: Repeatability, intermediate precision, reproducibility - Precision verification: Chi-square test against manufacturer claims - Precision profiles: CV vs concentration modeling with functional sensitivity
Quality Specifications - Biological variation-based goals: Calculate allowable total error from CVI and CVG - Sigma metrics: Six Sigma quality assessment - Performance assessment: Evaluate methods against quality specifications
All methods produce publication-ready plots and comprehensive statistical summaries.
library(valytics)
# Compare two creatinine measurement methods
data(creatinine_serum)
ba <- ba_analysis(
x = creatinine_serum$enzymatic,
y = creatinine_serum$jaffe
)
ba
#>
#> Bland-Altman Analysis
#> ----------------------------------------
#> n = 80 paired observations
#>
#> Difference type: Absolute (y - x)
#> Confidence level: 95%
#>
#> Results:
#> Bias (mean difference): 0.174
#> 95% CI: [0.127, 0.220]
#> SD of differences: 0.209
#>
#> Limits of Agreement:
#> Lower LoA: -0.236
#> 95% CI: [-0.316, -0.156]
#> Upper LoA: 0.584
#> 95% CI: [0.504, 0.663]plot(ba)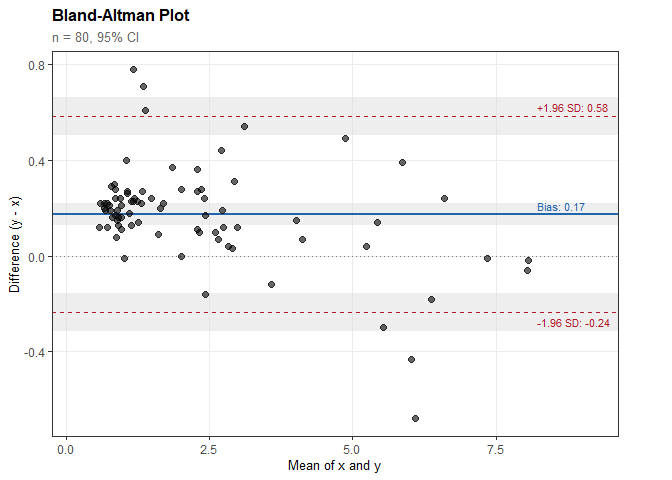
pb <- pb_regression(
x = creatinine_serum$enzymatic,
y = creatinine_serum$jaffe
)
pb
#>
#> Passing-Bablok Regression
#> ----------------------------------------
#> n = 80 paired observations
#>
#> CI method: Analytical (Passing-Bablok 1983)
#> Confidence level: 95%
#>
#> Regression equation:
#> creatinine_serum$jaffe = 0.234 + 0.971 * creatinine_serum$enzymatic
#>
#> Results:
#> Intercept: 0.234
#> 95% CI: [0.229, 0.239]
#> (excludes 0: significant constant bias)
#>
#> Slope: 0.971
#> 95% CI: [0.966, 0.974]
#> (excludes 1: significant proportional bias)plot(pb)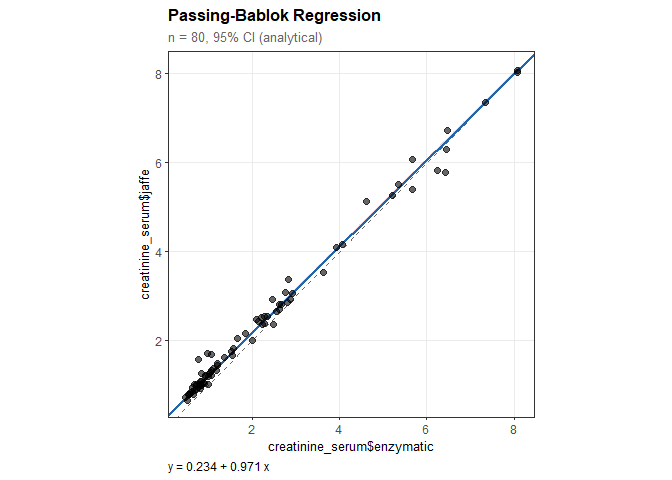
dm <- deming_regression(
x = creatinine_serum$enzymatic,
y = creatinine_serum$jaffe
)
dm
#>
#> Deming Regression
#> ----------------------------------------
#> n = 80 paired observations
#>
#> Error ratio (lambda): 1.000
#> CI method: Jackknife
#> Confidence level: 95%
#>
#> Regression equation:
#> creatinine_serum$jaffe = 0.277 + 0.953 * creatinine_serum$enzymatic
#>
#> Results:
#> Intercept: 0.277 (SE = 0.028)
#> 95% CI: [0.222, 0.332]
#> (excludes 0: significant constant bias)
#>
#> Slope: 0.953 (SE = 0.014)
#> 95% CI: [0.925, 0.982]
#> (excludes 1: significant proportional bias)plot(dm)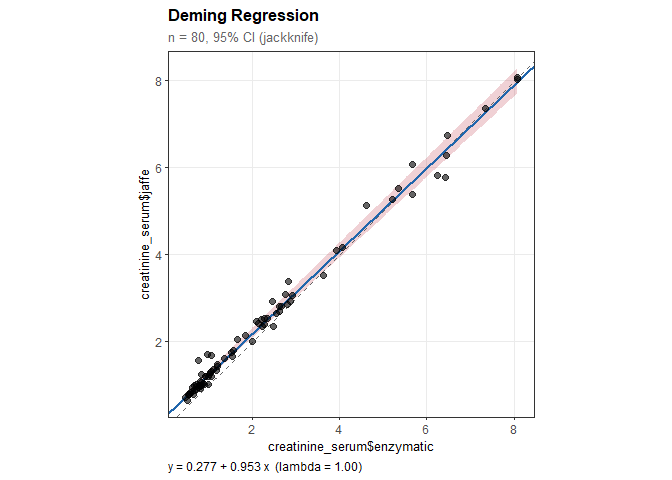
# Analyze precision from a multi-day, multi-run study
data(troponin_precision)
prec <- precision_study(
data = troponin_precision,
value = "value",
sample = "level",
day = "day",
run = "run"
)
prec
#>
#> Precision Study Analysis
#> ---------------------------------------------
#> n = 120 observations
#> Design: day/run/replicate (inferred)
#> Estimation: ANOVA (Method of Moments)
#> CI method: Satterthwaite, 95% CI
#>
#> Samples: 6 concentration levels
#> (Showing results for first sample; use $by_sample for all)
#>
#> Precision Estimates:
#> ---------------------------------------------
#> Repeatability: SD = 0.243 (CV = 4.86%)
#> 95% CI: [0.170, 0.426]
#> Between-run: SD = 0.188 (CV = 3.76%)
#> 95% CI: [0.117, 0.461]
#> Between-day: SD = 0.000 (CV = 0.00%)
#> 95% CI: [0.000, 0.000]
#> Within-laboratory precision: SD = 0.307 (CV = 6.15%)
#> 95% CI: [0.227, 0.476]plot(prec, type = "cv")
#> Warning in data.frame(sample = sample_names[i], mean_conc = if
#> (!is.null(sample_means)) sample_means[sample_names[i]] else i, : Zeilennamen
#> wurden in einer short Variablen gefunden und wurden verworfen
#> Warning in data.frame(sample = sample_names[i], mean_conc = if
#> (!is.null(sample_means)) sample_means[sample_names[i]] else i, : Zeilennamen
#> wurden in einer short Variablen gefunden und wurden verworfen
#> Warning in data.frame(sample = sample_names[i], mean_conc = if
#> (!is.null(sample_means)) sample_means[sample_names[i]] else i, : Zeilennamen
#> wurden in einer short Variablen gefunden und wurden verworfen
#> Warning in data.frame(sample = sample_names[i], mean_conc = if
#> (!is.null(sample_means)) sample_means[sample_names[i]] else i, : Zeilennamen
#> wurden in einer short Variablen gefunden und wurden verworfen
#> Warning in data.frame(sample = sample_names[i], mean_conc = if
#> (!is.null(sample_means)) sample_means[sample_names[i]] else i, : Zeilennamen
#> wurden in einer short Variablen gefunden und wurden verworfen
#> Warning in data.frame(sample = sample_names[i], mean_conc = if
#> (!is.null(sample_means)) sample_means[sample_names[i]] else i, : Zeilennamen
#> wurden in einer short Variablen gefunden und wurden verworfen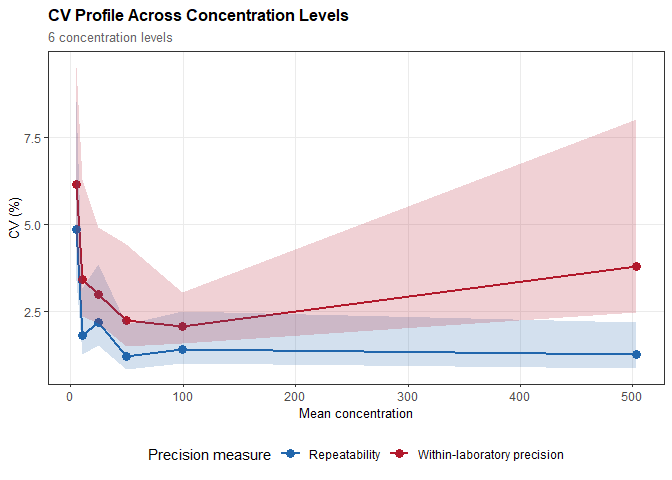
# Model CV vs concentration relationship
prof <- precision_profile(prec, cv_target = 10)
prof
#>
#> Precision Profile Analysis
#> ----------------------------------------
#> n = 6 concentration levels
#> Concentration range: 5 to 503 (100.75-fold)
#>
#> Model: Hyperbolic
#> CV = sqrt(2.667^2 + (26.905/x)^2)
#>
#> Parameters:
#> a = 2.667
#> b = 26.905
#>
#> Fit Quality:
#> R-squared = 0.816
#> RMSE = 0.578
#>
#> Functional Sensitivity:
#> CV = 10%: concentration = 2.792plot(prof)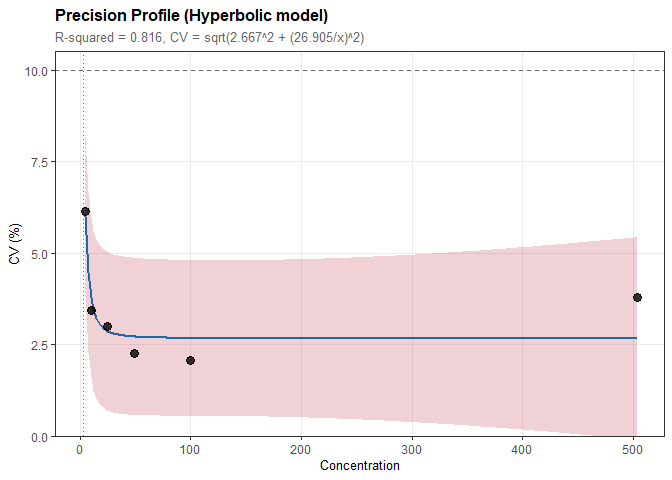
Calculate performance goals based on within-subject (CVI) and between-subject (CVG) biological variation:
# Creatinine biological variation (from EFLM database)
# CV_I = 5.95%, CV_G = 14.7%
ate <- ate_from_bv(cvi = 5.95, cvg = 14.7)
ate
#>
#> Analytical Performance Specifications from Biological Variation
#> ------------------------------------------------------------
#>
#> Input:
#> Within-subject CV (CV_I): 5.95%
#> Between-subject CV (CV_G): 14.70%
#> Performance level: desirable
#> Coverage factor (k): 1.65
#>
#> Specifications:
#> Allowable imprecision (CV_A): 2.98%
#> Allowable bias: 3.96%
#> Total allowable error (TEa): 8.87%# Evaluate method performance
sm <- sigma_metric(bias = 1.5, cv = 2.5, tea = 10)
sm
#>
#> Six Sigma Metric
#> ----------------------------------------
#>
#> Input:
#> Observed bias: 1.50%
#> Observed CV: 2.50%
#> Total allowable error (TEa): 10.00%
#>
#> Result:
#> Sigma: 3.40
#> Performance: Marginal
#> Defect rate: ~66,800 per million# Full quality assessment
assess <- ate_assessment(
bias = 1.5,
cv = 2.5,
tea = 10,
allowable_bias = 4.0,
allowable_cv = 3.0
)
assess
#>
#> Analytical Performance Assessment
#> --------------------------------------------------
#>
#> >>> METHOD ACCEPTABLE <<<
#>
#> Performance Summary:
#> Parameter Observed Allowable Status
#> -------------------- ---------- ---------- ----------
#> Bias 1.50% 4.00% PASS
#> CV (Imprecision) 2.50% 3.00% PASS
#> Total Error 5.62% 10.00% PASS
#>
#> Sigma Metric: 3.40 (Marginal)method1 ~ method2)The package includes four realistic clinical datasets:
| Dataset | Description | n |
|---|---|---|
glucose_methods |
POC meter vs. laboratory analyzer | 60 |
creatinine_serum |
Enzymatic vs. Jaffe methods | 80 |
troponin_cardiac |
Two hs-cTnI platforms | 50 |
troponin_precision |
hs-cTnI precision study (6 levels) | 120 |
Bland-Altman:
Passing-Bablok:
Deming:
Precision:
Biological Variation:
GPL-3