ggsurveillance is an R package with helpful tools and ggplot extensions for epidemiology, especially infectious disease surveillance and outbreak investigation. All functions provide tidy functional interfaces for easy integration with the tidyverse. For documentation and vignettes see: ggsurveillance.biostats.dev
geom_epicurve(): A ggplot geom for plotting epicurves.
stat_bin_date(): Date interval (week, month etc.) based
binning of case numbers with perfect alignment with e.g. reporting
week.geom_epicurve_text() and
geom_epicurve_point(): New geoms to easily add text
annotations or points to cases in epidemic curves.geom_vline_year(): Automatically detects the turn of
the year(s) from the date or datetime axis and draws a vertical
line.scale_y_cases_5er(): For better (case) count axis
breaks and positioning.bin_by_date(): A tidyverse-compatible
function for flexible date-based aggregation (binning).
align_dates_seasonal(): Align surveillance data for
seasonal plots (e.g. flu season).
geom_bar_diverging(): A geom for diverging bar charts,
which can be used to plot population pyramids, likert scales (sentiment
analyses) and other data with opposing categories, like vaccination
status or imported vs autochthonous (local) infections.
stat_diverging() for easy labeling of these charts with
category counts/percentages or total counts/percentagesscale_x_continuous_diverging() for symmetric diverging
scalesgeom_area_diverging() for continuous variables
(e.g. changes over time)geom_epigantt(): A geom for epigantt plots. Helpful to
visualize overlapping time intervals for contact tracing (e.g. hospital
outbreaks).
scale_y_discrete_reverse() which reverses the
order of the categorical scale.More ggplot2 add-ons:
guide_axis_nested_date(): An axis guide for creating
nested date labels for hierarchical time periods (e.g., year > month
> day).geom_label_last_value(): A geom for labeling the last
value of a time series (e.g. geom_line()).label_power10(): A ggplot2-compatible
labeling function to format numbers in scientific notation with powers
of 10 (e.g., \(2 \times 10^5\)).theme_mod_ functions for ggplot2 theme
modifications:
theme_mod_legend_position() etc. to adjust the legend
positions.theme_mod_rotate_x_axis_labels() etc. for rotating x
axis labels.theme_mod_remove_minor_grid() etc. to remove the minor
grid lines (x, y or both) or all grid lines.create_agegroups(): Create reproducible age groups
with highly customizable labels.
Additional utilities: geometric_mean(),
expand_counts(), and more
library(ggplot2)
library(tidyr)
library(outbreaks)
library(ggsurveillance)
sars_canada_2003 |> #SARS dataset from outbreaks
pivot_longer(starts_with("cases"),
names_prefix = "cases_",
names_to = "origin") |>
ggplot(aes(x = date, weight = value, fill = origin)) +
geom_epicurve(date_resolution = "week") +
geom_epicurve_text(aes(label = ifelse(origin == "travel", "🛪", "")),
date_resolution = "week", size = 1.5, color = "white") +
scale_x_date(date_labels = "W%V'%g", date_breaks = "2 weeks") +
scale_y_cases_5er() +
scale_fill_brewer(type = "qual", palette = 6) +
theme_classic()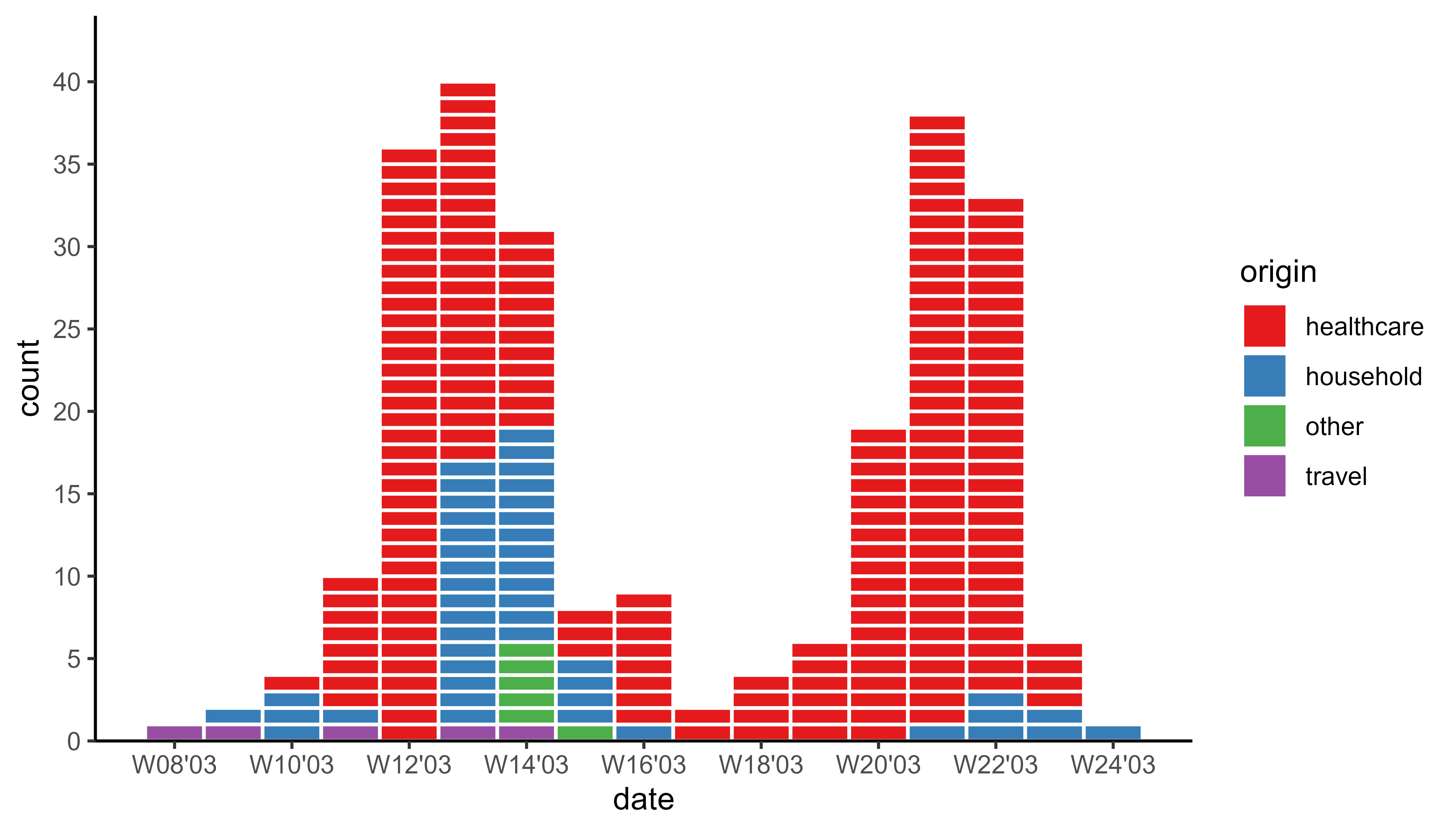
library(ggplot2)
library(dplyr)
library(ggsurveillance)
influenza_germany |>
filter(AgeGroup == "00+") |>
align_dates_seasonal(dates_from = ReportingWeek,
date_resolution = "isoweek",
start = 28) -> df_flu_aligned
ggplot(df_flu_aligned, aes(x = date_aligned, y = Incidence)) +
stat_summary(
aes(linetype = "Historical Median (Min-Max)"), data = . %>% filter(!current_season),
fun.data = median_hilow, geom = "ribbon", alpha = 0.3) +
stat_summary(
aes(linetype = "Historical Median (Min-Max)"), data = . %>% filter(!current_season),
fun = median, geom = "line") +
geom_line(
aes(linetype = "2024/25"), data = . %>% filter(current_season),
colour = "dodgerblue4", linewidth = 2) +
labs(linetype = NULL) +
scale_x_date(date_breaks = "month", date_labels = "%b'%Y",
guide = guide_axis_nested_date()) +
theme_bw() +
theme_mod_legend_position(position.inside = c(0.2, 0.8))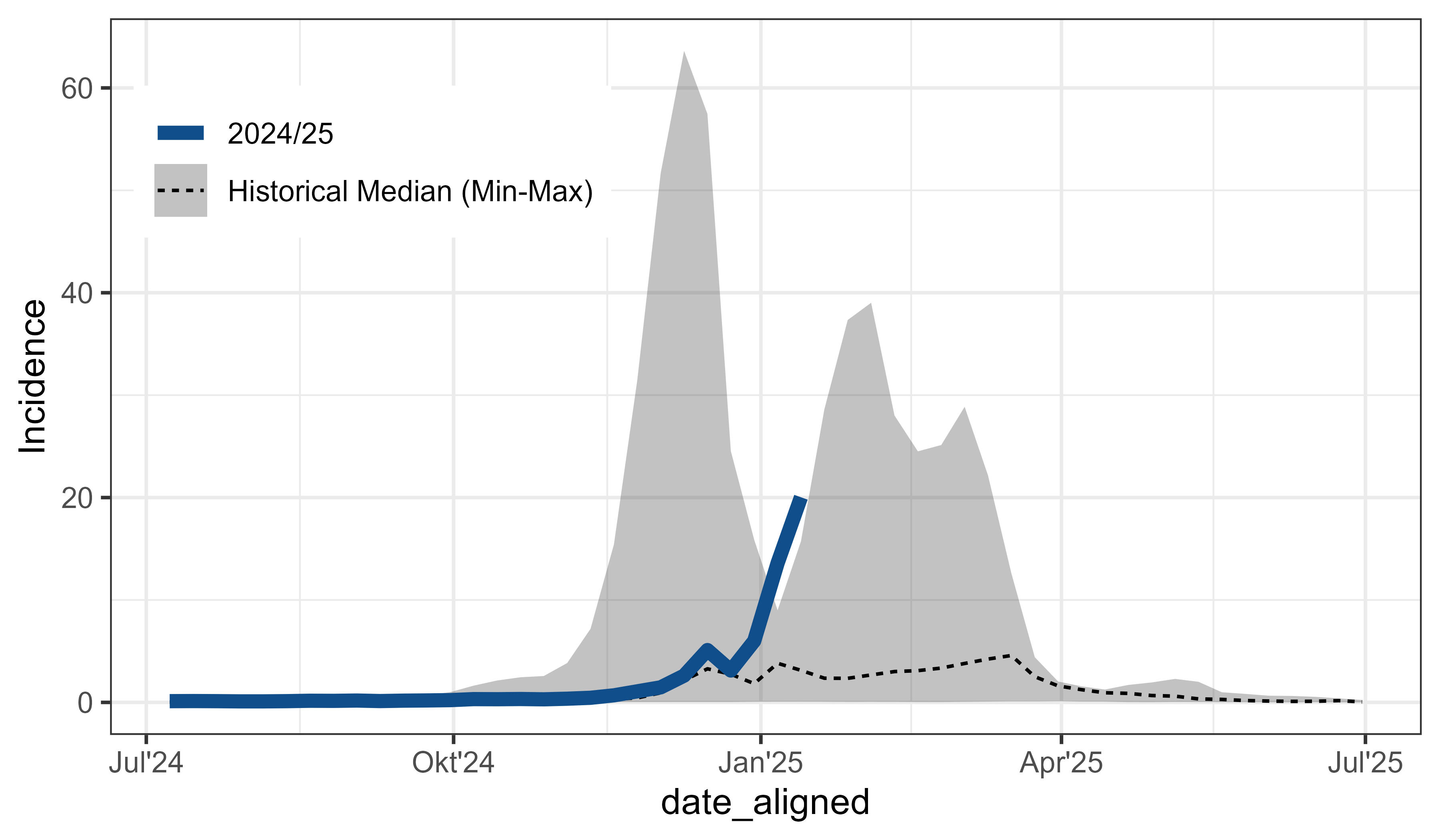
library(dplyr)
library(tidyr)
library(ggplot2)
library(ggsurveillance)
# Transform to long format
linelist_hospital_outbreak |>
pivot_longer(
cols = starts_with("ward"),
names_to = c(".value", "num"),
names_pattern = "ward_(name|start_of_stay|end_of_stay)_([0-9]+)",
values_drop_na = TRUE
) -> df_stays_long
linelist_hospital_outbreak |>
pivot_longer(cols = starts_with("pathogen"), values_to = "date") -> df_detections_long
# Plot
ggplot(df_stays_long) +
geom_epigantt(aes(y = Patient, xmin = start_of_stay, xmax = end_of_stay, color = name)) +
geom_point(aes(y = Patient, x = date, shape = "Date of pathogen detection"),
data = df_detections_long) +
scale_y_discrete_reverse() +
theme_bw() +
theme_mod_legend_bottom()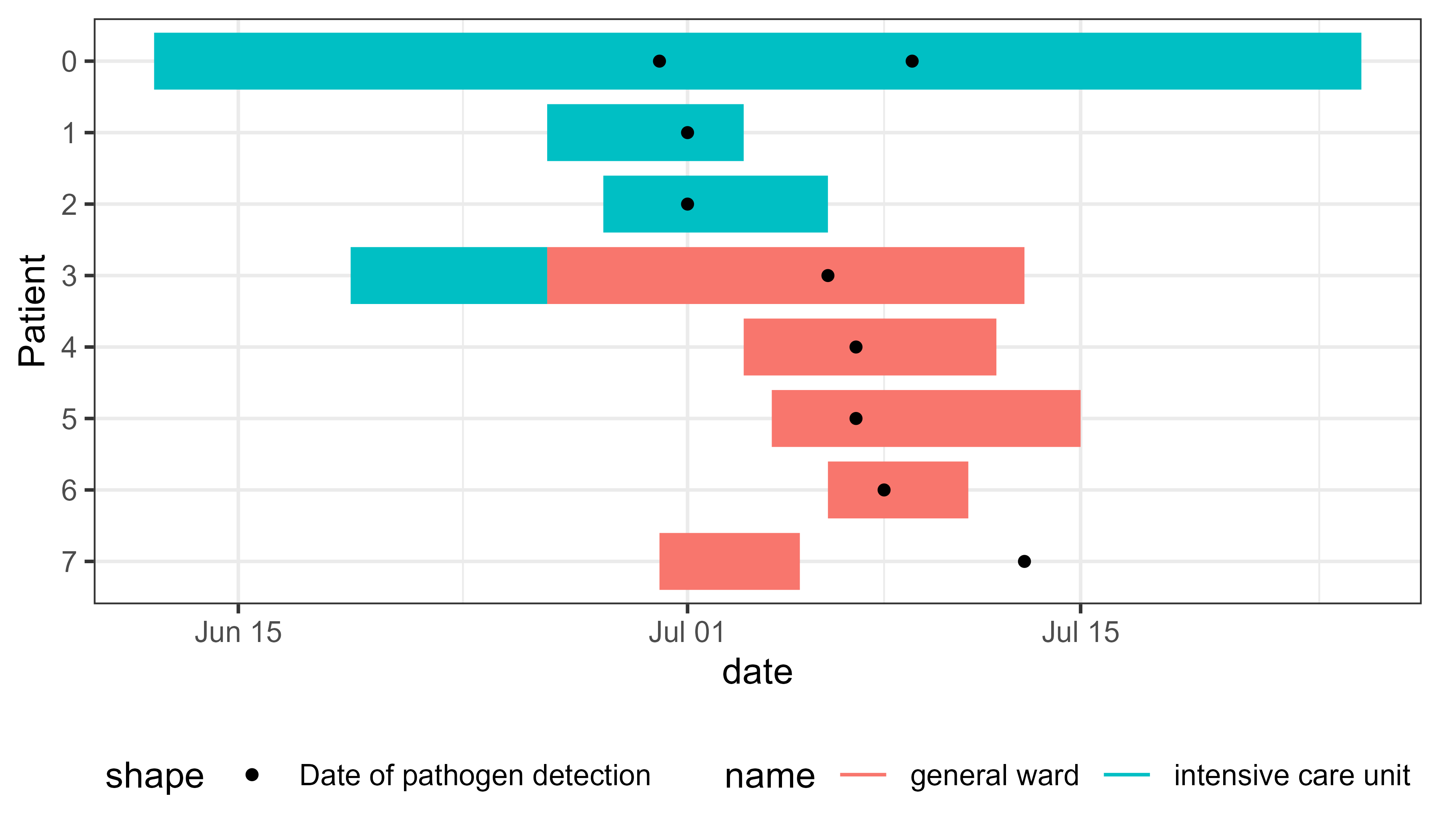
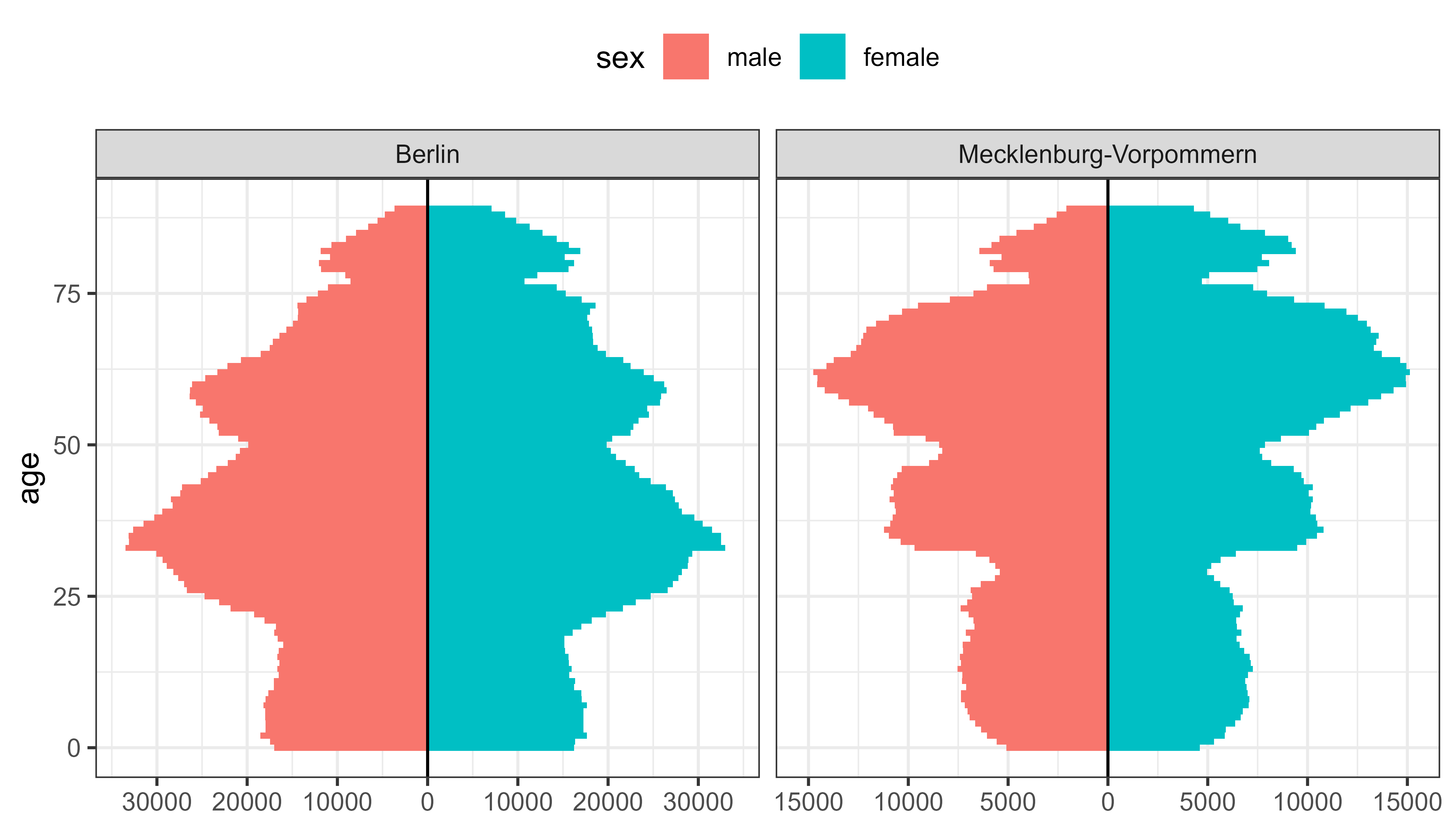
Useful for population pyramids, vaccination status, likert scales (sentiment) etc.
library(dplyr)
library(ggplot2)
library(ggsurveillance)
population_german_states |>
filter(state %in% c("Berlin", "Mecklenburg-Vorpommern"), age < 90) |>
ggplot(aes(y = age, fill = sex, weight = n)) +
geom_bar_diverging(width = 1) +
geom_vline(xintercept = 0) +
scale_x_continuous_diverging(n.breaks = 7) +
facet_wrap(~state, scales = "free_x") +
theme_bw() +
theme_mod_legend_top()