

Neuroimaging analyses produce region-level results – cortical thickness, p-values, network assignments – that need to end up on a brain figure. ggseg stores brain atlas geometries as simple features and plots them as ggplot2 layers, so you get publication-ready brain figures with the same code you’d use for any other ggplot.
Mowinckel & Vidal-Piñeiro (2020). Visualization of Brain Statistics With R Packages ggseg and ggseg3d. Advances in Methods and Practices in Psychological Science.
Install from CRAN:
install.packages("ggseg")Or get the development version from the ggsegverse r-universe:
options(repos = c(
ggsegverse = "https://ggsegverse.r-universe.dev",
CRAN = "https://cloud.r-project.org"
))
install.packages("ggseg")library(ggseg)
library(ggplot2)ggseg ships with three atlases: dk (Desikan-Killiany
cortical parcellation), aseg (automatic subcortical
segmentation), and tracula (white matter tracts).
plot() gives you a quick overview:
plot(dk())
plot(aseg())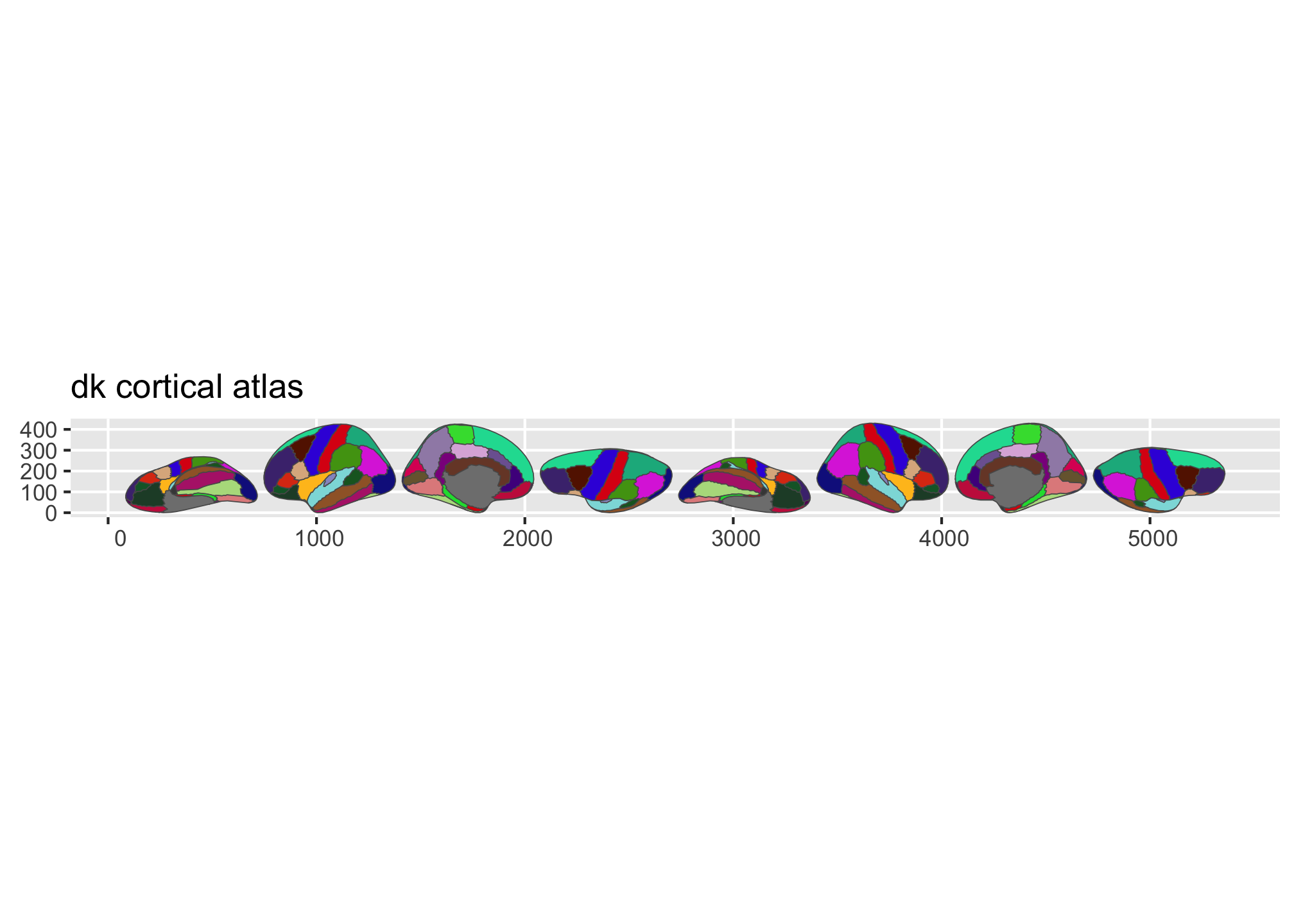
Figure 1: Overview of the dk and aseg built-in brain atlases.
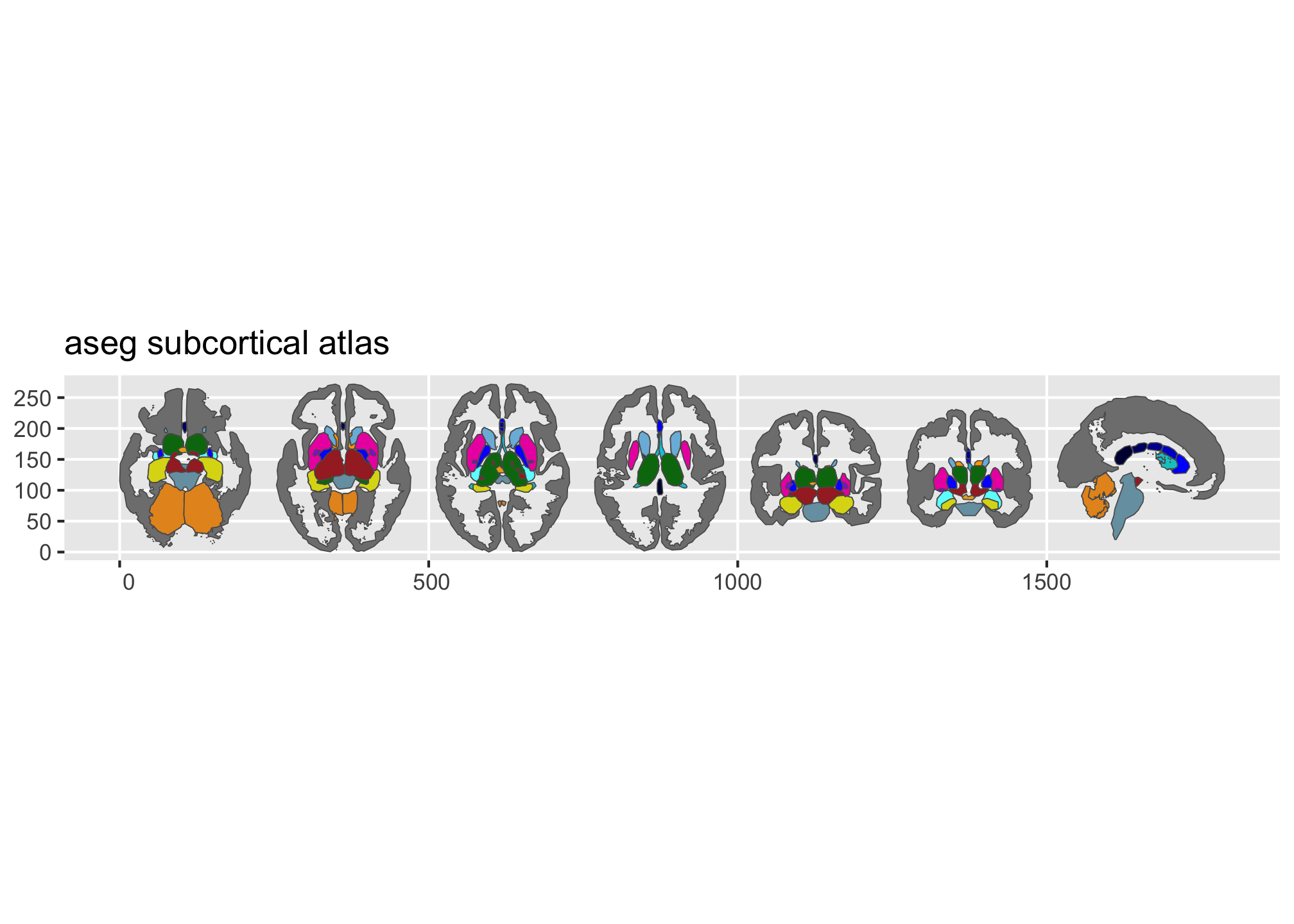
Figure 2: Overview of the dk and aseg built-in brain atlases.
Pass a data frame to ggplot() with a column that matches
the atlas (typically region or label).
geom_brain() handles the join:
library(dplyr)
some_data <- tibble(
region = rep(
c(
"transverse temporal",
"insula",
"precentral",
"superior parietal"
),
2
),
p = sample(seq(0, .5, .001), 8),
groups = c(rep("g1", 4), rep("g2", 4))
)
ggplot(some_data) +
geom_brain(
atlas = dk(),
position = position_brain(hemi ~ view),
aes(fill = p)
) +
facet_wrap(~groups) +
scale_fill_viridis_c(option = "cividis", direction = -1) +
theme_void()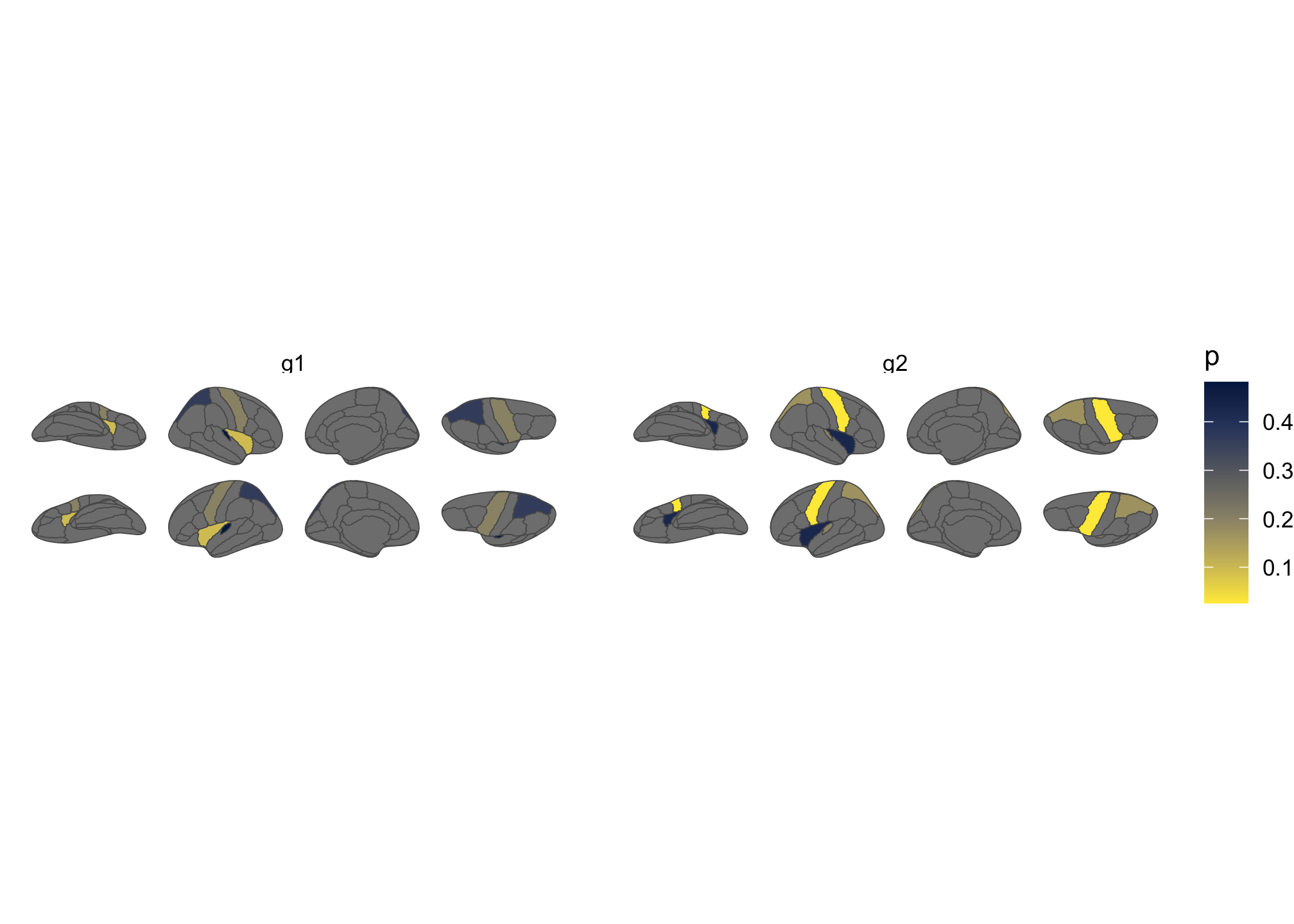
Figure 3: Brain plot coloured by external data, faceted by group.
Many additional atlases are available through the ggsegverse r-universe:
install.packages("ggsegYeo2011", repos = "https://ggsegverse.r-universe.dev")The package website
has vignettes covering external data, view positioning, the
geom_sf() workflow, and reading FreeSurfer stats files.
This tool is partly funded by:
EU Horizon 2020 Grant: Healthy minds 0-100 years: Optimising the use of European brain imaging cohorts (Lifebrain). Grant agreement number: 732592.