| Type: | Package |
| Title: | Retrieve and Analyze Clinical Trials Data from Public Registers |
| Version: | 1.26.0 |
| Date: | 2026-01-18 |
| Imports: | jsonlite, nodbi (≥ 0.13.1), rvest, stringdist, tidyr, V8, httr2, stringi, lubridate, jqr, dplyr, zip, readr, rlang, htmlwidgets |
| URL: | https://cran.r-project.org/package=ctrdata, https://rfhb.github.io/ctrdata/ |
| BugReports: | https://github.com/rfhb/ctrdata/issues |
| Description: | A system for querying, retrieving and analyzing protocol- and results-related information on clinical trials from four public registers, the 'European Union Clinical Trials Register' ('EUCTR', https://www.clinicaltrialsregister.eu/), 'ClinicalTrials.gov' (https://clinicaltrials.gov/ and also translating queries the retired classic interface), the 'ISRCTN' (http://www.isrctn.com/) and the 'European Union Clinical Trials Information System' ('CTIS', https://euclinicaltrials.eu/). Trial information is downloaded, converted and stored in a database ('PostgreSQL', 'SQLite', 'DuckDB' or 'MongoDB'; via package 'nodbi'). Protocols, statistical analysis plans, informed consent sheets and other documents in registers associated with trials can also be downloaded. Other functions implement trial concepts canonically across registers, identify deduplicated records, easily find and extract variables (fields) of interest even from complex nested data as used by the registers, merge variables and update queries. The package can be used for monitoring, meta- and trend-analysis of the design and conduct as well as of the results of clinical trials across registers. See overview in Herold, R. (2025) <doi:10.1017/rsm.2025.10061>. |
| License: | MIT + file LICENSE |
| RoxygenNote: | 7.3.3 |
| Suggests: | devtools, knitr, rmarkdown, RSQLite, mongolite, tinytest (≥ 1.2.1), RPostgres, duckdb, httr, tibble, clipr, chromote |
| VignetteBuilder: | knitr |
| NeedsCompilation: | no |
| Encoding: | UTF-8 |
| Language: | en-GB |
| Depends: | R (≥ 4.1) |
| Packaged: | 2026-01-18 08:18:20 UTC; ralfherold |
| Author: | Ralf Herold |
| Maintainer: | Ralf Herold <ralf.herold@mailbox.org> |
| Repository: | CRAN |
| Date/Publication: | 2026-01-18 08:30:02 UTC |
ctrdata: Retrieve and Analyze Clinical Trials Data from Public Registers
Description
A system for querying, retrieving and analyzing protocol- and results-related information on clinical trials from four public registers, the 'European Union Clinical Trials Register' ('EUCTR', https://www.clinicaltrialsregister.eu/), 'ClinicalTrials.gov' (https://clinicaltrials.gov/ and also translating queries the retired classic interface), the 'ISRCTN' (http://www.isrctn.com/) and the 'European Union Clinical Trials Information System' ('CTIS', https://euclinicaltrials.eu/). Trial information is downloaded, converted and stored in a database ('PostgreSQL', 'SQLite', 'DuckDB' or 'MongoDB'; via package 'nodbi'). Protocols, statistical analysis plans, informed consent sheets and other documents in registers associated with trials can also be downloaded. Other functions implement trial concepts canonically across registers, identify deduplicated records, easily find and extract variables (fields) of interest even from complex nested data as used by the registers, merge variables and update queries. The package can be used for monitoring, meta- and trend-analysis of the design and conduct as well as of the results of clinical trials across registers. See overview in Herold, R. (2025) doi:10.1017/rsm.2025.10061.
Author(s)
Maintainer: Ralf Herold ralf.herold@mailbox.org (ORCID)
Other contributors:
Marek Kubica (node-xml2js library) [copyright holder]
Ivan Bozhanov (jstree library) [copyright holder]
See Also
Useful links:
Report bugs at https://github.com/rfhb/ctrdata/issues
Check and prepare nodbi connection object for ctrdata
Description
Check and prepare nodbi connection object for ctrdata
Usage
ctrDb(con)
Arguments
con |
A database connection object, created with
|
Value
Connection object as list, with collection element under root
Find synonyms of an active substance
Description
An active substance can be identified by a recommended international nonproprietary name (INN), a trade or product name, or a company code(s). To find likely synonyms, the function retrieves from CTGOV2 the field protocolSection.armsInterventionsModule.interventions.otherNames. Note this does not seem to be based on choices from a dictionary but may be manually filled, thus is not free of error and needs to be checked.
Usage
ctrFindActiveSubstanceSynonyms(activesubstance = "", verbose = FALSE)
Arguments
activesubstance |
An active substance, in an atomic character vector |
verbose |
Print number of studies found in CTGOV2 for 'activesubstance' |
Value
A character vector of the active substance (input parameter) and synonyms, or NULL if active substance was not found and may be invalid
Examples
## Not run:
ctrFindActiveSubstanceSynonyms(activesubstance = "imatinib")
# [1] "imatinib" "CGP 57148" "CGP 57148B"
# [4] "CGP57148B" "Gleevec" "GLIVEC"
# [7] "Imatinib" "Imatinib Mesylate" "NSC 716051"
# [10] "ST1571" "STI 571" "STI571"
## End(Not run)
Generates queries that work across registers
Description
From high-level search terms provided by the user, generate specific queries for each registers with which ctrdata works, see ctrdata-registers. Search terms that are expanded to concepts such as from MeSH and MedDRA by the search implementations in registers include the 'intervention' and 'condition'. Logical operators only work with 'searchPhrase'.
Usage
ctrGenerateQueries(
searchPhrase = NULL,
condition = NULL,
intervention = NULL,
phase = NULL,
population = NULL,
recruitment = NULL,
startBefore = NULL,
startAfter = NULL,
completedBefore = NULL,
completedAfter = NULL,
onlyMedIntervTrials = TRUE,
onlyWithResults = FALSE,
countries = NULL
)
Arguments
searchPhrase |
String with optional logical operators ("AND", "OR") that will be searched in selected fields of registers that can handle logical operators (general or title fields), should not include quotation marks |
condition |
String with condition / disease |
intervention |
String with intervention |
phase |
String, e.g. "phase 2" (note that "phase 2+3" is a specific category, not the union set of "phase 2" and "phase 3") |
population |
String, e.g. "P" (paediatric), "A" (adult), "P+A" (adult and paediatric), "E" (elderly), "P+A+E" participants can be recruited. For ISRCTN, works only for "P", "A" or "E" but not mixed populations. |
recruitment |
String, one of "ongoing", "completed", "other" ( which includes "ended early" but this cannot be searched; use trial concept f.statusRecruitment to identify this status) |
startBefore |
String that can be interpreted as date (for EUCTR, when trial was first registered) |
startAfter |
String that can be interpreted as date (for EUCTR, when trial was first registered) |
completedBefore |
String that can be interpreted as date (does not work with EUCTR) |
completedAfter |
String that can be interpreted as date (does not work with EUCTR) |
onlyMedIntervTrials |
Logical, default |
onlyWithResults |
Logical |
countries |
Vector of country names, two- or three-letter ISO 3166 codes |
Value
Named vector of URLs for finding trials in the registers and as input to functions ctrLoadQueryIntoDb and ctrOpenSearchPagesInBrowser
Examples
urls <- ctrGenerateQueries(
intervention = "antibody",
phase = "phase 3",
startAfter = "2000-01-01")
# open queries in register web interface
sapply(urls, ctrOpenSearchPagesInBrowser)
# For CTIS to accept such a search URL and show results, consider installing
# the Tampermonkey browser extension from https://www.tampermonkey.net/,
# click on the extension icon, "Create a new script", "Utility" and then
# "Import from this URL":
# https://raw.githubusercontent.com/rfhb/ctrdata/master/tools/ctrdataURLcopier.js
# find potential research platform and platform trials
urls <- ctrGenerateQueries(
searchPhrase = paste0(
"basket OR platform OR umbrella OR master protocol OR ",
"multiarm OR multistage OR subprotocol OR substudy OR ",
"multi-arm OR multi-stage OR sub-protocol OR sub-study"),
phase = "phase 3",
startAfter = "01/31/2010",
countries = c("DE", "US", "United Kingdom"))
# open queries in register web interface
sapply(urls, ctrOpenSearchPagesInBrowser)
urls <- ctrGenerateQueries(
searchPhrase = "antibody AND covid",
recruitment = "completed",
)
## Not run:
# count trials found
sapply(urls, ctrLoadQueryIntoDb, only.count = TRUE)
# load queries into database collection
dbc <- nodbi::src_sqlite(collection = "my_collection")
sapply(urls, ctrLoadQueryIntoDb, con = dbc, euctrprotocolsall = FALSE)
## End(Not run)
Get register name and query parameters from search URL
Description
Extracts query parameters and register name from parameter 'url' or from the clipboard, into which the URL of a register search was copied.
Usage
ctrGetQueryUrl(url = "", register = "")
Arguments
url |
URL such as from the browser address bar. If not specified, clipboard contents will be checked for a suitable URL. For automatically copying the user's query of a register in a web browser to the clipboard, see here. Can also contain a query term such as from dbQueryHistory()["query-term"]. Can also be an identifier of a trial, which based on its format will indicate to which register it relates. |
register |
Optional name of register (one of "EUCTR", "CTGOV2" "ISRCTN" or "CTIS") in case 'url' is a query term but not a full URL |
Details
To obtain the search query from CTIS, consider installing the Tampermonkey browser extension, click on the extension icon, "Create a new script", "Utility" and then "Import from this URL": 'https://raw.githubusercontent.com/rfhb/ctrdata/master/tools/ctrdataURLcopier.js' This will copy query URLs from all registers into the clipboard; see ctrOpenSearchPagesInBrowser for additional uses.
Value
A data frame (or tibble, if tibble is loaded)
with column names 'query-term' and 'query-register'.
The data frame (or tibble) can be passed as such as parameter
'queryterm' to ctrLoadQueryIntoDb and as parameter
'url' to ctrOpenSearchPagesInBrowser.
Examples
# user copied into the clipboard the URL from
# the address bar of the browser that shows results
# from a query in one of the trial registers
if (interactive()) try(ctrGetQueryUrl(), silent = TRUE)
# extract query parameters from search result URL
# (URL was cut for the purpose of formatting only)
ctrGetQueryUrl(
url = paste0(
"https://classic.clinicaltrials.gov/ct2/results?",
"cond=&term=AREA%5BMaximumAge%5D+RANGE%5B0+days%2C+28+days%5D",
"&type=Intr&rslt=&age_v=&gndr=&intr=Drugs%2C+Investigational",
"&titles=&outc=&spons=&lead=&id=&cntry=&state=&city=&dist=",
"&locn=&phase=2&rsub=&strd_s=01%2F01%2F2015&strd_e=01%2F01%2F2016",
"&prcd_s=&prcd_e=&sfpd_s=&sfpd_e=&rfpd_s=&rfpd_e=&lupd_s=&lupd_e=&sort="
)
)
# other examples
ctrGetQueryUrl("https://www.clinicaltrialsregister.eu/ctr-search/trial/2007-000371-42/results")
ctrGetQueryUrl("https://euclinicaltrials.eu/ctis-public/view/2022-500041-24-00")
ctrGetQueryUrl("https://classic.clinicaltrials.gov/ct2/show/NCT01492673?cond=neuroblastoma")
ctrGetQueryUrl("https://clinicaltrials.gov/ct2/show/NCT01492673?cond=neuroblastoma")
ctrGetQueryUrl("https://clinicaltrials.gov/study/NCT01467986?aggFilters=ages:child")
ctrGetQueryUrl("https://www.isrctn.com/ISRCTN70039829")
# using identifiers of single trials
ctrGetQueryUrl("70039829")
ctrGetQueryUrl("ISRCTN70039829")
ctrGetQueryUrl("NCT00617929")
ctrGetQueryUrl("2022-501142-30-00")
ctrGetQueryUrl("2012-003632-23")
Load and store register trial information
Description
Retrieves information on clinical trials from registers and stores it in a collection in a database. Main function of ctrdata for accessing registers. A collection can store trial information from different queries or different registers. When at least one trial is loaded, query details are stored in the collection and can be accessed using dbQueryHistory. A previous query can be re-run, which replaces existing or adds new trial records while keeping any user annotations of existing trial records.
Usage
ctrLoadQueryIntoDb(
queryterm = NULL,
register = "",
querytoupdate = NULL,
forcetoupdate = FALSE,
euctrresults = FALSE,
euctrresultshistory = FALSE,
euctrprotocolsall = TRUE,
ctgov2history = FALSE,
ctishistory = FALSE,
documents.path = NULL,
documents.regexp = "prot|sample|statist|sap_|p1ar|p2ars|icf|ctalett|lay|^[0-9]+ ",
annotation.text = "",
annotation.mode = "append",
only.count = FALSE,
con = NULL,
verbose = FALSE
)
Arguments
queryterm |
Either a single string with the full URL of a search
query in a register, e.g. one of the strings returned by
ctrGenerateQueries, or a single-row data frame returned by
ctrGetQueryUrl or dbQueryHistory,
or an '_id' in the format of one of the trial registers (e.g., "NCT..."),
or, together with |
register |
String with abbreviation of register to query,
either "EUCTR", "CTGOV2", "ISRCTN" or "CTIS". Not needed
if |
querytoupdate |
Either the word "last", or the row number of
a query in the data frame returned by dbQueryHistory that
should be run to retrieve any new or update trial records since
this query was run the last time.
This parameter takes precedence over |
forcetoupdate |
If |
euctrresults |
If |
euctrresultshistory |
If |
euctrprotocolsall |
If |
ctgov2history |
For trials from CTGOV2, retrieve historic
versions of the record. Default is |
ctishistory |
If |
documents.path |
If this is a relative or absolute
path to a directory that exists or can be created,
save any documents into it that are directly available from
the register ("EUCTR", "CTGOV2", "ISRCTN", "CTIS")
such as PDFs on results, analysis plans, spreadsheets,
patient information sheets, assessments or product information.
Default is |
documents.regexp |
Regular expression, case insensitive,
to select documents by filename, if saving documents is requested
(see |
annotation.text |
Text to be including into the field
"annotation" in the records retrieved with the query
that is to be loaded into the collection.
The contents of the field "annotation" for a trial record
are preserved e.g. when running this function again and
loading a record of a with an annotation, see parameter
|
annotation.mode |
One of "append" (default), "prepend" or "replace" for new annotation.text with respect to any existing annotation for the records retrieved with the query that is to be loaded into the collection. |
only.count |
Set to |
con |
A database connection object, created with
|
verbose |
If |
Value
A list with elements 'n' (number of trial records newly imported or updated), ‘success' (a vector of _id’s of successfully loaded records), 'failed' (a vector of identifiers of records that failed to load) and 'queryterm' (the query term used). The returned list has several attributes (including database and collection name, as well as the query history of this database collection) to facilitate documentation.
Examples
## Not run:
# Count ongoing interventional cancer trials involving children
# Note this query is a classical CTGOV query and is translated
# to a corresponding query for the current CTGOV2 webinterface
ctrLoadQueryIntoDb(
queryterm = "cond=cancer&recr=Open&type=Intr&age=0",
register = "CTGOV",
only.count = TRUE
)
dbc <- nodbi::src_sqlite(collection = "my_collection")
# Retrieve protocol- and results-related information
# on two specific trials identified by their EU number,
# including protocol versions from all Member States
ctrLoadQueryIntoDb(
queryterm = "2005-001267-63+OR+2008-003606-33",
register = "EUCTR",
euctrresults = TRUE,
euctrprotocolsall = TRUE,
con = dbc
)
# Retrieve all information on more than 40 trials
# that are labelled as phase 3 and that mention
# either neuroblastoma or lymphoma from ISRCTN,
# into the same collection as used before
ctrLoadQueryIntoDb(
queryterm = paste0(
"https://www.isrctn.com/search?",
"q=neuroblastoma+OR+lymphoma&filters=phase%3APhase+III"),
con = dbc
)
# Retrieve information trials in CTIS mentioning neonates
ctrLoadQueryIntoDb(
queryterm = paste0("https://euclinicaltrials.eu/ctis-public/",
"search#searchCriteria={%22containAll%22:%22%22,",
"%22containAny%22:%22neonates%22,%22containNot%22:%22%22}"),
con = dbc
)
## End(Not run)
Open register to show query results or search page
Description
Open advanced search pages of register(s), or execute search in browser. For CTIS to accept a search URL and show results, consider installing the Tampermonkey browser extension, click on the extension icon, "Create a new script", "Utility" and then "Import from this URL": 'https://raw.githubusercontent.com/rfhb/ctrdata/master/tools/ctrdataURLcopier.js'
Usage
ctrOpenSearchPagesInBrowser(url = "", register = "", copyright = FALSE)
Arguments
url |
of search results page to show in the browser. To open the
browser with a previous search, the output of ctrGetQueryUrl
or dbQueryHistory can be used. Can be left as empty string
(default) to open the advanced search page of |
register |
Register(s) to open, "EUCTR", "CTGOV2", "ISRCTN" or "CTIS". Default is empty string, and this opens the advanced search page of the registers (including the expert search page in the case of CTGOV). |
copyright |
(Optional) If set to |
Value
(String) Full URL corresponding to the shortened url
in conjunction with register if any, or invisibly
TRUE if no url is specified.
Examples
# Open all and check copyrights before using registers
ctrOpenSearchPagesInBrowser(copyright = TRUE)
# Open specific register advanced search page
ctrOpenSearchPagesInBrowser(register = "CTGOV2")
ctrOpenSearchPagesInBrowser(register = "CTIS")
ctrOpenSearchPagesInBrowser(register = "EUCTR")
ctrOpenSearchPagesInBrowser(register = "ISRCTN")
# Open all queries that were loaded into demo collection
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials",
flags = RSQLite::SQLITE_RO)
dbh <- dbQueryHistory(con = dbc)
for (r in seq_len(nrow(dbh))) {
ctrOpenSearchPagesInBrowser(dbh[r, ])
}
Show full structure and all data of a trial
Description
If used interactively, the function shows a widget of all data in the trial
as a tree of field names and values. The widget opens in the default browser.
Fields names and values can be search and selected. Selected fields can be
copied to the clipboard for use with function dbGetFieldsIntoDf.
The trial is retrieved with ctrLoadQueryIntoDb if no database
con is provided or if the trial is not in database con.
For use in a Shiny app, see output and render functions in source code
here.
Usage
ctrShowOneTrial(identifier = NULL, con = NULL)
Arguments
identifier |
A trial identifier ('_id') string, see examples |
con |
A database connection object, created with
|
Details
This is the widget for CTIS trial 2022-501142-30-00:
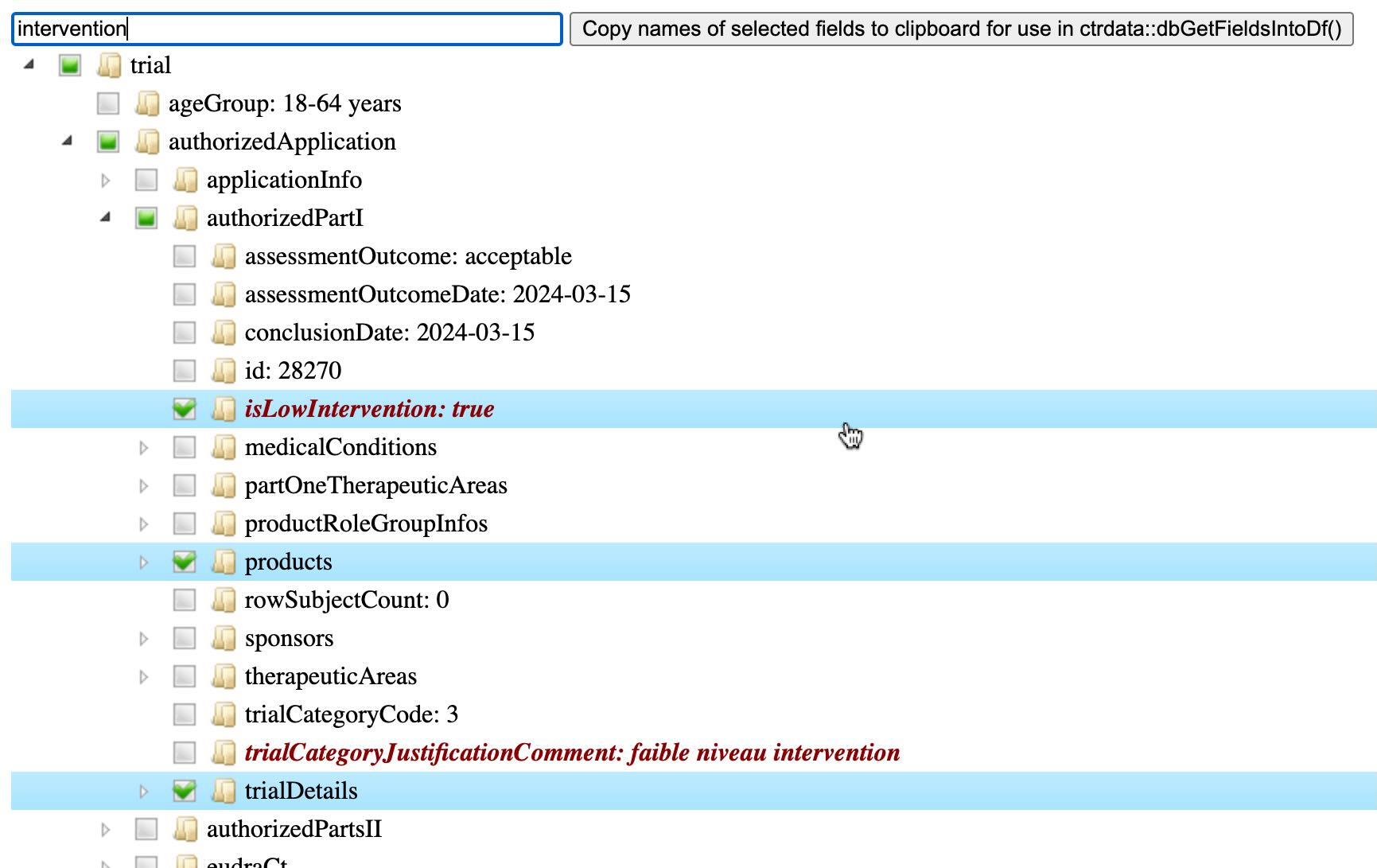
Value
Invisibly, the trial data for constructing an HTML widget.
Examples
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials",
flags = RSQLite::SQLITE_RO)
# get sample of identifiers of trials in database
sample(dbFindIdsUniqueTrials(con = dbc), 5L)
# all such identifiers work
id <- "2014-003556-31"
id <- "2014-003556-31-SE"
id <- "76463425"
id <- "ISRCTN76463425"
id <- "NCT03431558"
id <- "2022-501142-30-00"
# note these ids also work with
# ctrGetQueryUrl(url = id) and
# ctrLoadQueryIntoDb(queryterm = id, ...)
# show widget for user to explore and search content as well as to
# select fields of interest and to click on "Copy names of selected
# fields to clipboard..." to use them with dbGetFieldsIntoDf()
ctrShowOneTrial(identifier = id, con = dbc)
Getting started, database connection, function overview
Description
ctrdata is a package for aggregating and analysing data on clinical
studies, and for obtaining documents, from public trial registers
1 - Define a database connection
Package ctrdata retrieves trial data and stores it in a database
collection. The connection is specified using nodbi, which allows to
use different database backends in an identical way.
A database connection object is specified once and then can be used as
parameter con in subsequent calls of ctrdata functions.
Specifying collection = "<my trial data collection's name>"
indicates the table in the database that package ctrdata should use.
| Database | Connection object |
| SQLite | dbc <- nodbi::src_sqlite(dbname = "my_db", collection = "my_coll") |
| DuckDB | dbc <- nodbi::src_duckdb(dbname = "my_db", collection = "my_coll") |
| MongoDB | dbc <- nodbi::src_mongo(db = "my_db", collection = "my_coll") |
| PostgreSQL | dbc <- nodbi::src_postgres(dbname = "my_db"); dbc[["collection"]] <- "my_coll"
|
2 - Load information from clinical trial registers
ctrGenerateQueries (generate from simple user input specific queries for registers EUCTR, CTIS, CTGOV2 and ISRCTN), ctrOpenSearchPagesInBrowser (open queries in browser), see script (automatically copy user search in any register to clipboard), see ctrdata-registers for details on registers and how to search, ctrLoadQueryIntoDb (load trial records found with query into database collection).
3 - Use database with downloaded trial information
ctrShowOneTrial (show widget to explore structure, fields and data of a trial), dbFindFields (find names of fields of interest in trial records in a collection), dbGetFieldsIntoDf (create a data frame with fields of interest and calculated trial concepts from collection), ctrdata-trial-concepts (calculate pre-defined trial concepts across registers), dbFindIdsUniqueTrials (get de-duplicated identifiers of clinical trials' records to subset a data frame).
4 - Operate on a trial data frame from dbGetFieldsIntoDf
dfTrials2Long (convert fields with nested elements into long format), dfName2Value (get values for variable(s) of interest), ctrdata-trial-concepts (calculate pre-defined trial concepts for every register).
Author(s)
Ralf Herold ralf.herold@mailbox.org
See Also
Useful links:
Report bugs at https://github.com/rfhb/ctrdata/issues
Information on clinical trial registers
Description
Registers of the four clinical trial registers from which package ctrdata can retrieve, aggregate and analyse protocol- and result-related information as well as documents, last updated 2026-01-11.
1 - Overview
-
EUCTR: The EU Clinical Trials Register is complete with 44,366 clinical trials (at least one investigational medicinal product, IMP; in the European Union and beyond), including more than 25,800 trials with results, which continue to be added.
-
CTIS: The EU Clinical Trials Information System, launched in 2023, holds more than 10,680 publicly accessible clinical trials, including more than 520 with results or a report. (To automatically get CTIS search query URLs, see here)
-
CTGOV2: ClinicalTrials.gov holds more than 565,000 interventional and observational studies, including more than 71,000 interventional studies with results.
-
ISRCTN: The ISRCTN Registry holds more than 27,500 interventional and observational health studies, including almost 15,000 studies with results.
| CTGOV2 | CTIS | EUCTR | ISRCTN | |
| Protocol-related information | Structured | Structured | Structured | Structured |
| Result-related information | Structured, publication links | Documents | Structured, documents | Publication links |
| Documents | Primarily protocol, SAP | Protocol, info sheets, SAP, results | Reports | Protocol, info sheets, SAP, results |
2 - Notable changes
CTGOV "classic" was retired on 2024-06-25; ctrdata subsequently translates
CTGOV queries to CTGOV2 queries. The new website ("CTGOV2") can be used with
ctrdata since 2023-08-27. Database collections created with CTGOV queries
can still be used since functions in ctrdata continue to support them.
CTIS was relaunched on 2024-06-17, changing the data structure and search
syntax, to which ctrdata was updated.
CTIS can be used with ctrdata since 2023-03-25.
EUCTR removed search parameter status= as of February 2025.
More information on changes:
here.
3 - References
| Material | EUCTR | CTGOV2 | ISRCTN | CTIS |
| About | link | link | link | link |
| Terms & conditions, disclaimer | link | link | link | link |
| How to search | link | link | link | link |
| Search interface | link | link | link | link |
| Expert / advanced search | link | link | link | link |
| Glossary / related information | link | link | link | link |
| FAQ / caveats / examples | link | link, link, link | link | link |
| Data dictionaries / structure | link, link, link | link, link, link | link | link (see XLSX files) |
| Example* (see below) | link | link | link | link |
Some registers are expanding entered search terms using dictionaries (example).
4 - Example and ctrdata motivation
See vignette ctrdata_summarise for several other examples.
*This example is an expert search for interventional trials primarily with neonates, investigating treatments for infectious conditions. It shows that searches in the web interface of most registers are not sufficient to identify the trials of interest:
EUCTR retrieves trials with neonates, but not only those exclusively in neonates.
ISRCTN retrieves studies with interventions other than medicines.
CTIS retrieves trials that mention the words neonates and infection. (To show CTIS search results, see here)
To address this issue, trials can be retrieved with ctrLoadQueryIntoDb into a database collection and in a second step trials of interest can be selected based on values of relevant fields, for example:
EUCTR field
f115_children_211yearsand other age group criteriaISRCTN field
interventions.intervention.interventionTypefor type of studyCTIS fields
ageGroupandauthorizedApplication.authorizedPartI.medicalConditions.medicalCondition
ctrdata supports users with pre-defined ctrdata-trial-concepts and
these cover the example above, and with functions dbFindFields and
ctrShowOneTrial for finding fields of interest and reviewing data
structure, respectively.
Author(s)
Ralf Herold ralf.herold@mailbox.org
Trial concepts implemented across registers
Description
ctrdata includes (since version 1.21.0) functions that implement selected
trial concepts. Concepts of clinical trials, such as their start or status of
recruitment, require to analyse several fields against various
pre-defined values. The structure and value sets of fields differ between
all ctrdata-registers. In this situation, the implemented trial
concepts simplify and accelerate a user's analysis workflow and also increase
analysis consistency.
Details
The implementation of trial concepts in ctrdata has not been validated
with any formal approach, but has been checked for plausibility and
against expectations. The implementation is based on current
understanding, on public data models and on scientific papers, as relevant.
As with other R functions, call help("f.startDate") or print its
implementation code by entering the name of the function as command,
e.g. f.startDate.
Please raise an issue here
to ask about or improve a trial concept.
The following trial concepts can be used by referencing their name when
calling dbGetFieldsIntoDf (parameter calculate).
Concepts will continue to be refined and added;
last updated 2026-01-11.
-
f.assignmentType (factor) was the assignment to treatment based on randomisation or not? ("R" or "NR")
-
f.controlType (factor) which type of internal or concurrent control is used in the trial? ("none", "no-treatment", "placebo", "active", "placebo+active" or "other")
-
f.externalLinks (character) provides links to publications or other external references
-
f.hasResults (logical) are any types of results recorded, e.g., structured data, reports or publications
-
f.isMedIntervTrial (logical) is the trial interventional and does it have one or more medicines (drugs or biological) as investigational (experimental) intervention? (irrespective of status of authorisation and of study design)
-
f.isUniqueTrial (logical) is the trial record unique in the data frame of trial records, based on default parameters of dbFindIdsUniqueTrials?
-
f.likelyPlatformTrial (logical, list of likely related trials, and list of maybe related trials) is the trial possibly a (research) platform trial, and what are related trials? (based on trial title,
f.numTestArmsSubstances, number of periods; identifiers of related trials; similarity of terms in parts of trial titles) -
f.numSites (integer) how many sites does the trial have?
-
f.numTestArmsSubstances (integer) how many arms or groups in the trial have medicines that are investigational? (cannot be calculated for ISRCTN or for phase 1 trials)
-
f.primaryEndpointDescription (list of character) string containing protocol definition, details and time frames, concatenated with " == "
-
f.primaryEndpointResults (columns of number, character, integer) returning the statistical testing p value and method as well as the number of subjects included in the test, each in one new column, for the first primary endpoint only
-
f.resultsDate (date) the planned or achieved date of results availability
-
f.startDate (date) the planned, authorised or documented date of start of recruitment
-
f.sampleSize (integer) the planned or achieved number of subjects or participants recruited (depending on status of recruitment and availability of results; for "CTIS" always planned number)
-
f.sponsorType (factor) a type or class of sponsor(s) that is simplified to "not for profit", "for profit", "mixed" or "other"
-
f.statusRecruitment (factor) a status that is simplified to "ongoing" (includes temporarily halted), "completed", "ended early" (includes terminated or ended prematurely) and "other" (includes planned, not yet recruiting, stopped, withdrawn)
-
f.trialObjectives (string) identifies with letters those objectives that could be identified in text fragments, e.g. "E S PD D", with "E" (efficacy), "S" (safety), "D" (dose-finding)
-
f.trialPhase (ordered factor) the phase(s) of medicine development with which a trial is associated ("phase 1", "phase 1+2" etc.)
-
f.trialPopulation (columns of factor, string and string) age groups (e.g., "P" for paediatric participants, "A" for adults, "E" for older than 65 years, or "P+A"), inclusion and exclusion criteria texts
-
f.trialTitle (string) full or scientific title of the study
Author(s)
Ralf Herold ralf.herold@mailbox.org
Find names of fields in the database collection
Description
Given a part of the name of fields that are of interest to the user, this
function returns the full field names used in records that were
previously loaded into a collection (using ctrLoadQueryIntoDb).
Only names of fields that have a value in the collection can be returned.
Set sample = FALSE to force screening all records in the
collection for field names, which can take long.
See ctrShowOneTrial to interactively find fields.
Usage
dbFindFields(namepart = ".*", con, sample = TRUE, verbose = FALSE)
Arguments
namepart |
A character string (can be a regular expression, including Perl-style) to be searched among all field names (keys) in the collection, case-insensitive. The default '".*"' lists all fields. |
con |
A database connection object, created with
|
sample |
If |
verbose |
If |
Details
The full names of child fields are returned in dot notation (e.g.,
clinical_results.outcome_list.outcome.measure.class_list.class.title)
In addition, names of parent fields (e.g.,
clinical_results) are returned.
Data in parent fields is typically complex (nested), see
dfTrials2Long for easily handling it.
For field definitions of the registers, see
"Definition" in ctrdata-registers.
Note: When dbFindFields is first called after
ctrLoadQueryIntoDb, it will take a moment.
Value
Vector of strings with full names of field(s) found, ordered by register and alphabet, see examples. Names of the vector are the names of the register holding the respective fields. The field names can be fed into dbGetFieldsIntoDf to extract the data for the field(s) from the collection into a data frame.
Examples
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials",
flags = RSQLite::SQLITE_RO)
dbFindFields(namepart = "date", con = dbc)[1:5]
# view names of all 3838 fields from all registers:
allFields <- dbFindFields(con = dbc, sample = FALSE)
if (interactive()) View(data.frame(
register = names(allFields),
field = allFields))
Get identifiers of deduplicated trial records
Description
Records for a clinical trial can be loaded from more than one register into a collection. This function returns deduplicated identifiers for all trials in the collection, respecting the register(s) preferred by the user. All registers are recording identifiers also from other registers, which are used by this function to provide a vector of identifiers of deduplicated trials.
Usage
dbFindIdsUniqueTrials(
preferregister = c("CTGOV2", "EUCTR", "CTGOV", "ISRCTN", "CTIS"),
prefermemberstate = "BE",
include3rdcountrytrials = TRUE,
con,
verbose = FALSE
)
Arguments
preferregister |
A vector of the order of preference for
registers from which to generate unique _id's, default
|
prefermemberstate |
Code of single EU Member State for which records
should returned. If not available, a record for BE or lacking this, any
random Member State's record for the trial will be returned.
For a list of codes of EU Member States, please see vector
|
include3rdcountrytrials |
A logical value if trials should be retained
that are conducted exclusively in third countries, that is, outside
the European Union. Ignored if |
con |
A database connection object, created with
|
verbose |
If |
Details
Note that the content of records may differ between registers (and, for "EUCTR", between records for different Member States). Such differences are not considered by this function.
Note that for "CTIS", the trial with the latest (highest) resubmission number (last two digits of 'id', clinical trial number) is identified.
Note that the trial concept f.isUniqueTrial (which uses this function) can be calculated at the time of creating a data frame with dbGetFieldsIntoDf, which often may be the preferred approach.
Value
A named vector with strings of keys (field "_id") of records in the collection that represent unique trials, where names correspond to the register of the record.
Examples
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials",
flags = RSQLite::SQLITE_RO)
dbFindIdsUniqueTrials(con = dbc)[1:10]
# alternative as of ctrdata version 1.21.0,
# using defaults of dbFindIdsUniqueTrials()
df <- dbGetFieldsIntoDf(
fields = "keyword",
calculate = "f.isUniqueTrial",
con = dbc)
# using base R
df[df[[".isUniqueTrial"]], ]
## Not run:
library(dplyr)
df %>% filter(.isUniqueTrial)
## End(Not run)
Create data frame of specified fields or trial concepts from database collection
Description
Fields in the collection are retrieved from all records into a data frame (or tibble). The function uses the field names to appropriately type the values that it returns, harmonising original values (e.g., "Yes" to 'TRUE', "false" to 'FALSE', "Information not present in EudraCT" to 'NA', date strings to dates or time differences, number strings to numbers). Trial concepts are calculated for all records and included in the return value.
Usage
dbGetFieldsIntoDf(fields = "", calculate = "", con, verbose = FALSE)
Arguments
fields |
Vector of one or more strings, with names of sought fields. See function dbFindFields for how to find names of fields and ctrShowOneTrial for interactively selecting field names. Dot path notation ("field.subfield") without indices is supported. If compatibility with nodbi::src_postgres is needed, specify fewer than 50 fields, or use parent fields such as '"a.b"' instead of 'c("a.b.c.d", "a.b.c.e")' and then access sought fields with dfTrials2Long followed by dfName2Value or with other R functions. |
calculate |
Vector of one or more strings, which are names of functions to calculate certain trial concepts from fields in the collection, across different registers. See ctrdata-trial-concepts for available functions. |
con |
A database connection object, created with
|
verbose |
If |
Details
Within a given trial record, a field can be hierarchical and structured, that is, nested. The function simplifies the structure of nested data and may concatenate multiple strings in a field using " / " (see example) and may have widened the returned data frame with additional columns that were recursively expanded from simply nested data (e.g., "externalRefs" to columns "externalRefs.doi", "externalRefs.eudraCTNumber" etc.). For an alternative ways for handling complex nested data, see dfTrials2Long and dfName2Value for extracting the sought variable(s).
Value
A data frame (or tibble, if tibble is loaded)
with columns corresponding to the sought fields.
A column with the record '_id' will always be included.
The maximum number of rows of the returned data frame is equal to the number
of trial records in the database collection, or less if none of the fields
has a value in a record.
Examples
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials",
flags = RSQLite::SQLITE_RO)
# get fields that are nested within another field
# and can have multiple values with the nested field
dbGetFieldsIntoDf(
fields = "b1_sponsor.b31_and_b32_status_of_the_sponsor",
con = dbc)
# fields that are lists of string values are
# returned by concatenating values with " / "
dbGetFieldsIntoDf(
fields = "keyword",
con = dbc)
# calculate new field(s) from data across trials
df <- dbGetFieldsIntoDf(
fields = "keyword",
calculate = c("f.statusRecruitment", "f.isUniqueTrial", "f.startDate"),
con = dbc)
table(df$.statusRecruitment, exclude = NULL)
## Not run:
library(dplyr)
library(ggplot2)
df %>%
filter(.isUniqueTrial) %>%
count(.statusRecruitment)
df %>%
filter(.isUniqueTrial) %>%
ggplot() +
stat_ecdf(aes(
x = .startDate,
colour = .statusRecruitment))
## End(Not run)
Show history of queries loaded into collection
Description
Note that a new entry is added to the history of queries when ctrLoadQueryIntoDb retrieves at least 1 clinical trial.
Usage
dbQueryHistory(con, verbose = FALSE)
Arguments
con |
A database connection object, created with
|
verbose |
If |
Value
A data frame (or tibble, if tibble is loaded)
with columns: 'query-timestamp', 'query-register',
'query-records' (note: this is the number of records loaded when last
executing ctrLoadQueryIntoDb, not the total record number) and
'query-term', with one row for each time that
ctrLoadQueryIntoDb loaded trial records into this collection.
Examples
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials",
flags = RSQLite::SQLITE_RO)
dbQueryHistory(con = dbc)
Merge variables, keeping type where possible, optionally relevel factors
Description
Merge variables in a data frame such as returned by dbGetFieldsIntoDf into a new variable, and optionally also map its values to new levels. See ctrdata-trial-concepts for pre-defined cross-register concepts that are already implemented based on merging fields from different registers and calculating a new field.
Usage
dfMergeVariablesRelevel(df = NULL, colnames = "", levelslist = NULL)
Arguments
df |
A data.frame with the variables (columns) to be merged into one vector. |
colnames |
A vector of names of columns in 'df' that hold the variables
to be merged, or a selection of columns as per |
levelslist |
A names list with one slice each for a new value to be used for a vector of old values (optional). |
Value
A vector, with the type of the columns to be merged
Examples
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials",
flags = RSQLite::SQLITE_RO)
df <- dbGetFieldsIntoDf(
fields = c(
"ctrname",
"protocolSection.eligibilityModule.healthyVolunteers",
"f31_healthy_volunteers",
"eligibility.healthy_volunteers",
"participants.participantType",
paste0(
"authorizedPartI.trialDetails.trialInformation.",
"populationOfTrialSubjects.clinicalTrialGroups.name"
),
paste0(
"authorizedApplication.authorizedPartI.trialDetails.trialInformation.",
"populationOfTrialSubjects.clinicalTrialGroups.name"
)
),
con = dbc
)
df$isrctnhealthy <- "Healthy volunteer" == df[, 6]
df$ctis1healthy <- "Healthy volunteers" == df[ ,7]
df$ctis2healthy <- "Healthy volunteers" == df[, 8]
table(
df$ctrname,
dfMergeVariablesRelevel(
df = df,
colnames = 'matches("healthy")'
))
Get value for variable of interest
Description
Get information for variable of interest (e.g., clinical endpoints) from a long data frame of protocol- or result-related trial information as returned by dfTrials2Long. Parameters 'valuename', 'wherename' and 'wherevalue' are matched using Perl regular expressions and ignoring case. See also ctrdata-trial-concepts for how to extract and calculate concepts of interest across registers.
Usage
dfName2Value(df, valuename = "", wherename = "", wherevalue = "")
Arguments
df |
A data frame (or tibble) with four columns ('_id', 'identifier', 'name', 'value') as returned by dfTrials2Long |
valuename |
A character string for the name of the field that holds the value of the variable of interest (e.g., a summary measure such as "endPoints.*tendencyValue.value") |
wherename |
(optional) A character string to identify the variable of interest among those that repeatedly occur in a trial record (e.g., "endPoints.endPoint.title") |
wherevalue |
(optional) A character string with the value of the variable identified by 'wherename' (e.g., "response") |
Value
A data frame (or tibble, if tibble is loaded)
that includes the values of interest, with columns
'_id', 'identifier', 'name', 'value' and 'where' (with the
contents of 'wherevalue' found at 'wherename').
Contents of 'value' are strings unless all its elements
are numbers. The 'identifier' is generated by
function dfTrials2Long to identify matching elements,
e.g endpoint descriptions and measurements.
Examples
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials",
flags = RSQLite::SQLITE_RO)
dfwide <- dbGetFieldsIntoDf(
fields = c(
## ctgov - typical results fields
# "clinical_results.baseline.analyzed_list.analyzed.count_list.count",
# "clinical_results.baseline.group_list.group",
# "clinical_results.baseline.analyzed_list.analyzed.units",
"clinical_results.outcome_list.outcome",
"study_design_info.allocation",
## euctr - typical results fields
# "trialInformation.fullTitle",
# "baselineCharacteristics.baselineReportingGroups.baselineReportingGroup",
# "trialChanges.hasGlobalInterruptions",
# "subjectAnalysisSets",
# "adverseEvents.seriousAdverseEvents.seriousAdverseEvent",
"endPoints.endPoint",
"subjectDisposition.recruitmentDetails"
), con = dbc
)
dflong <- dfTrials2Long(df = dfwide)
## get values for the endpoint 'response'
dfName2Value(
df = dflong,
valuename = paste0(
"clinical_results.*measurement.value|",
"clinical_results.*outcome.measure.units|",
"endPoints.endPoint.*tendencyValue.value|",
"endPoints.endPoint.unit"
),
wherename = paste0(
"clinical_results.*outcome.measure.title|",
"endPoints.endPoint.title"
),
wherevalue = "response"
)
Convert data frame with trial records into long format
Description
The function works with procotol- and results- related information.
It converts lists and other values that are in a data frame returned
by dbGetFieldsIntoDf into individual rows of a long data frame.
From the resulting long data frame, values of interest can be selected
using dfName2Value.
The function is particularly useful for fields with complex content,
such as node field "clinical_results" from EUCTR, for which
dbGetFieldsIntoDf returns as a multiply nested list and for
which this function then converts every observation of every (leaf)
field into a row of its own.
Usage
dfTrials2Long(df)
Arguments
df |
Data frame (or tibble) with columns including
the trial identifier ( |
Value
A data frame (or tibble, if tibble is loaded)
with the four columns: '_id', 'identifier', 'name', 'value'
Examples
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials",
flags = RSQLite::SQLITE_RO)
dfwide <- dbGetFieldsIntoDf(
fields = "clinical_results.participant_flow",
con = dbc)
dfTrials2Long(df = dfwide)
Calculate type of assignment to intervention in a study
Description
Calculate type of assignment to intervention in a study
Usage
f.assignmentType(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.assignmentType', which is a factor with levels 'R' (randomised assignment) and 'NR' (all other types of assignment).
Examples
# fields needed
f.assignmentType()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
field = "ctrname",
calculate = "f.assignmentType",
con = dbc)
trialsDf
Calculate type of control data collected in a study
Description
Trial concept calculated: type of internal control. ICH E10 lists as types of control: placebo concurrent control, no-treatment concurrent control, dose-response concurrent control, active (positive) concurrent control, external (including historical) control, multiple control groups. Dose-controlled trials are currently not identified. External (including historical) controls are so far not identified in specific register fields. Cross-over designs, where identifiable, have active controls.
Usage
f.controlType(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.controlType', which is a factor with levels 'none', 'no-treatment', 'placebo', 'active', 'placebo+active' and 'other'.
Examples
# fields needed
f.controlType()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
field = "ctrname",
calculate = "f.controlType",
con = dbc)
trialsDf
Calculate the external references from a study's register record
Description
Trial concept calculated: Calculates the links e.g. to publications or other external files referenced from a study record. Requires loading results-related information for EUCTR. Note that documents stored in registers can be downloaded directly, see ctrLoadQueryIntoDb.
Usage
f.externalLinks(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and new column '.externalLinks' (character).
Examples
# fields needed
f.hasResults()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.externalLinks",
con = dbc)
trialsDf
Calculate if a study's results are available
Description
Trial concept calculated: Calculates if results have been recorded in the register, as structured data, reports or publications, for example. Requires loading results-related information for EUCTR.
Usage
f.hasResults(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and new column '.hasResults' (logical).
Examples
# fields needed
f.hasResults()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.hasResults",
con = dbc)
trialsDf
Calculate if study is a medicine-interventional study
Description
Trial concept calculated: Calculates if record is a medicine-interventional trial, investigating one or more medicine, whether biological or not. For EUCTR and CTIS, this corresponds to all records as per the definition of the EU Clinical Trial Regulation. For CTGOV and CTGOV2, this is based on drug or biological as type of intervention, and interventional as type of study. For ISRCTN, this is based on drug or biological as type of intervention, and interventional as type of study.
Usage
f.isMedIntervTrial(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with colums '_id' and '.isMedIntervTrial', a logical.
Examples
# fields needed
f.isMedIntervTrial()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.isMedIntervTrial",
con = dbc)
trialsDf
Calculate if record is unique for a study
Description
Trial concept calculated: Applies function dbFindIdsUniqueTrials() with its defaults.
Usage
f.isUniqueTrial(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.isUniqueTrial', a logical.
Examples
# fields needed
f.isUniqueTrial()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
fields = "ctrname",
calculate = "f.isUniqueTrial",
con = dbc)
trialsDf
Calculate if study is likely a platform trial or not
Description
Trial concept calculated: platform trial, research platform. As operational definition, at least one of these criteria is true: a. trial has "platform", "basket", "umbrella", "multi.?arm", "multi.?stage" or "master protocol" in its title or description (for ISRCTN, this is the only criterion; some trials in EUCTR lack data in English), b. trial has more than 2 active arms with different investigational medicines, after excluding comparator, auxiliary and placebo medicines (calculated with f.numTestArmsSubstances; not used for ISRCTN because it cannot be calculated precisely), c. trial has more than 2 periods, after excluding safety run-in, screening, enrolling, extension and follow-up periods (for CTGOV and CTGOV2, this criterion requires results-related data).
Usage
f.likelyPlatformTrial(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Details
For EUCTR, requires that results have been included in the collection, using 'ctrLoadQueryIntoDb(queryterm = ..., euctrresults = TRUE, con = ...)'. Requires packages dplyr and stringdist to be installed; stringdist is used for evaluating terms in brackets in the trial title, where trials may be related if the term similarity is 0.77 or higher.
Publication references considered: EU-PEARL WP2 2020 and Williams RJ et al. 2022, doi:10.1136/bmj-2021-067745
Value
data frame with columns '_id' and '.likelyPlatformTrial', a logical, and two complementary columns, each with lists of identifiers: '.likelyRelatedTrials' (based on other identifiers provided in the trial record, including 'associatedClinicalTrials' from CTIS; listing identifiers whether or not the trial with the other identifier is in the database collection) and '.maybeRelatedTrials' (based on similar short terms in the first set of brackets or before a colon in the trial title; only listing identifiers from the database collection).
Examples
# fields needed
f.likelyPlatformTrial()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.likelyPlatformTrial",
con = dbc)
trialsDf
Calculate number of sites of a study
Description
Trial concept calculated: number of the sites where the trial is conducted. EUCTR lacks information on number of sites outside of the EEA; for each non-EEA country mentioned, at least one site is assumed.
Usage
f.numSites(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.numSites', an integer.
Examples
# fields needed
f.numSites()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.numSites",
con = dbc)
trialsDf
Calculate number of arms or groups with investigational medicines in a study
Description
Trial concept calculated: number of active arms with different investigational medicines, after excluding non-active comparator, auxiliary and placebo arms / medicines. For ISRCTN, this is imprecise because arms are not identified in a field. Most registers provide no or only limited information on phase 1 trials, so that this number typically cannot be calculated for these trials. Requires packages stringdist to be installed; stringdist is used for evaluating names of active substances, which are considered similar when the similarity is 0.8 or higher.
Usage
f.numTestArmsSubstances(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.numTestArmsSubstances', an integer
Examples
# fields needed
f.numTestArmsSubstances()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.numTestArmsSubstances",
con = dbc)
trialsDf
Calculate details of a primary endpoint of a study
Description
Trial concept calculated: full description of the primary endpoint, concatenating with " == " its title, description, time frame of assessment. The details vary by register. The text description can be used for identifying trials of interest or for analysing trends in primary endpoints, which among the set of all endpoints are most often used for determining the number of participants sought for the study.
Usage
f.primaryEndpointDescription(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.primaryEndpointDescription', which is a list (that is, one or more items in one vector per row; the background is that some trials have several endpoints as primary).
Examples
# fields needed
f.primaryEndpointDescription()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.primaryEndpointDescription",
con = dbc)
trialsDf
Calculate details of a study's primary endpoint analysis and testing
Description
Trial concept calculated: Calculates several results-related elements of the primary analysis of the primary endpoint. Requires loading results-related information. For CTIS and ISRCTN, such information is not available in structured format. Recommended to be combined with .controlType, .sampleSize etc. for analyses.
Usage
f.primaryEndpointResults(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and new columns: '.primaryEndpointFirstPvalue' (discarding any inequality indicator, e.g. <=), '.primaryEndpointFirstPmethod' (normalised string, e.g. chisquared), '.primaryEndpointFirstPsize' (number included in test, across assignment groups).
Examples
# fields needed
f.primaryEndpointResults()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.primaryEndpointResults",
con = dbc)
trialsDf
Calculate date of results of a study
Description
Trial concept calculated: earliest date of results as recorded in the register. At that date, results may have been incomplete and may have been changed later. For EUCTR, requires that results and preferrably also their history of publication have been included in the collection, using ctrLoadQueryIntoDb(queryterm = ..., euctrresultshistory = TRUE, con = ...). Cannot be calculated for ISRCTN, which does not have a corresponding field.
Usage
f.resultsDate(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.resultsDate', a date.
Examples
# fields needed
f.resultsDate()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.resultsDate",
con = dbc)
trialsDf
Calculate sample size of a study
Description
Trial concept calculated: sample size of the trial, preferring results-related (achieved recruitment) over protocol-related information (planned sample size). Thus, the calculated number depends on the status of the recruitment (see f.statusRecruitment) and on the availability of results data; for "CTIS", the number always corresponds to the planned sample size.
Usage
f.sampleSize(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.sampleSize', an integer.
Examples
# fields needed
f.sampleSize()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.sampleSize",
con = dbc)
trialsDf
Calculate type of sponsor of a study
Description
Trial concept calculated: type or class of the sponsor(s) of the study. No specific field is available in ISRCTN; thus, sponsor type is set to 'other'. Note: If several sponsors, sponsor type is deemed 'mixed' if there is both, a commercial and a non-commercial sponsor(s).
Usage
f.sponsorType(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.sponsorType', which is a factor with levels 'for profit', 'not for profit', 'mixed' (not and for profit sponsors) or 'other'.
Examples
# fields needed
f.sponsorType()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.sponsorType",
con = dbc)
trialsDf
Calculate start date of a study
Description
Trial concept calculated: start of the trial, based on the documented or planned start of recruitment, or on the date of opinion of the competent authority.
Usage
f.startDate(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.startDate', a date.
Examples
# fields needed
f.startDate()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
field = "ctrname",
calculate = "f.startDate",
con = dbc)
trialsDf
Calculate status of recruitment of a study
Description
Trial concept calculated: status of recruitment at the time of loading the trial records. Maps the categories that are in fields which specify the state of recruitment. Simplifies the status into three categories.
Usage
f.statusRecruitment(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.statusRecruitment', which is a factor with levels 'ongoing' (includes active, not yet recruiting; temporarily halted; suspended; authorised, not started and similar), 'completed' (includes ended; ongoing, recruitment ended), 'ended early' (includes prematurely ended, terminated early) and 'other' (includes revoked, withdrawn, planned, stopped).
Examples
# fields needed
f.statusRecruitment()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.statusRecruitment",
con = dbc)
trialsDf
Calculate objectives of a study
Description
Trial concept calculated: objectives of the trial, by searching for text fragments found in fields describing its purpose, objective, background or hypothesis, after applying .isMedIntervTrial, because the text fragments are tailored to medicinal product interventional trials. This is a simplification, and it is expected that the criteria will be further refined. The text fragments only apply to English.
Usage
f.trialObjectives(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.trialObjectives', which is a string with letters separated by a space, such as E (efficacy, including cure, survival, effectiveness); A (activity, including reponse, remission, seroconversion); S (safety); PK; PD (including biomarker); D (dose-finding, determining recommended dose); LT (long-term); and FU (follow-up).
Examples
# fields needed
f.trialObjectives()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.trialObjectives",
con = dbc)
trialsDf
Calculate phase of a clinical trial
Description
Trial concept calculated: phase of a clinical trial as per ICH E8(R1).
Usage
f.trialPhase(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.trialPhase', which is an ordered factor with levels 'phase 1', 'phase 1+2', 'phase 2', 'phase 2+3', 'phase 2+4', 'phase 3', 'phase 3+4', 'phase 1+2+3', 'phase 4', 'phase 1+2+3+4'.
Examples
# fields needed
f.trialPhase()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.trialPhase",
con = dbc)
trialsDf
Calculate in- and exclusion criteria and age groups
Description
Trial concept calculated: inclusion and exclusion criteria as well as age groups that can participate in a trial, based on protocol-related information. Since CTGOV uses single text field for eligibility criteria, text extraction is used to separate in- and exclusion criteria. (See dfMergeVariablesRelevel with an example for healthy volunteers.)
Usage
f.trialPopulation(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and new columns: '.trialPopulationAgeGroup' (factor, "P", "A", "P+A", "E", "A+E", "P+A+E"), '.trialPopulationInclusion' (string), '.trialPopulationExclusion' (string).
Examples
# fields needed
f.trialPopulation()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.trialPopulation",
con = dbc)
trialsDf
Calculate the title of a study
Description
Trial concept calculated: scientific or full title of the study.
Usage
f.trialTitle(df = NULL)
Arguments
df |
data frame such as from dbGetFieldsIntoDf. If 'NULL', prints fields needed in 'df' for calculating this trial concept, which can be used with dbGetFieldsIntoDf. |
Value
data frame with columns '_id' and '.trialTitle', a string.
Examples
# fields needed
f.trialTitle()
# apply trial concept when creating data frame
dbc <- nodbi::src_sqlite(
dbname = system.file("extdata", "demo.sqlite", package = "ctrdata"),
collection = "my_trials", flags = RSQLite::SQLITE_RO)
trialsDf <- dbGetFieldsIntoDf(
calculate = "f.trialTitle",
con = dbc)
trialsDf